First Human chromosome Y sequence was published nearly two decades ago, yet the full chr Y sequence is not competently resolved due to high repeat content. Chr Y is a highly complex and posses great challenges in generating a high quality full length assembly. Human chr Y measure about ~57mb in length. Male-specific region of the Y chromosome, MSY, differentiates the sexes and comprises 95% of the chromosome’s length. MSY is a mosaic of hetero-chromatic sequences with three euchromatic classes: X-transposed, X-degenerate and ampliconic. These classes contain all 156 known transcription units, which include 78 protein-coding genes that collectively encode 27 distinct proteins.
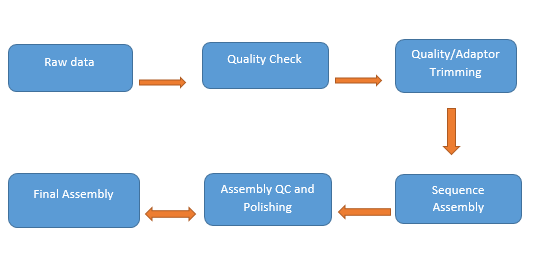
We here make an attempt to assembly human Chr Y with publicly available chromosome Y raw data sequenced on Minion in fast5 file format from EBI under study accession PRJEB28143. Raw starting coverage is ~44x coverage, while after correction step of Canu v1.8 we got ~30x coverage as input for sequence assembly. Canu generated a draft assembly of size ~23.5 mb with 166 contigs. This draft assembly was then polished using Oxford Nanopore raw reads with Racon followed by a short read polishing with Pilon. Final assembly quality was accessed using Quast, dnadiff (Mummer) and alignment against reference genome GRCh38 using LAST. Contigs that mapped elsewhere on human reference genome with a % higher base mapping as compared to chromosome Y were filtered out. Next a gap filling step was run with dnaasm package using raw Oxford Nanopore long reads and repeat masker for repeat annotation.
Oxford Nanopore Assembly Work Flow
Assembly Stats Table
| Assembly | No. of Contigs | Largest Contigs (bps) | N50 (bps) | Size (Mb) | Percent Identity* |
| Raw Assembly | 166 | 1,428,304 | 451,553 | 23.568 | 97.89% |
| Polished Assembly | 128 | 1,668,953 | 486,086 | 23.676 | 99.23% |
| Final Assembly | 80 | 1,668,953 | 553,052 | 21.404 | 99.57% |
* % identity to Y chromosome aligned bases.
We also compared our assembly with PacBio assembly of the Ashkenazim Trio sample from “Genome in a Bottle” consortium. NA24385 had a 30x sequencing depth on chr Y. These Pacbio long reads were assembled using Canu v1.6, draft assembly comprised of 193 contigs , with an N50 of 213 kb, covering 15.3 Mb. Draft assembly was aligned to reference genome Grch38 using Last (v.984). Contigs were further filtered using lastal package and alignments having mismap probability higher than 10e-5.
Comparison of Chromosome Y Assembly stats:
| Assembly | No. Of contigs | N50 | Size |
| Nanopore Long Reads | 80 | ~553kb | 21.4mb |
| Pacbio Long Reads | 184 | ~215kb | 15.3mb |
Clearly, we generated a more contigous Chr Y assembly using Oxford Nanopore reads compared to the published Pacbio assembly. Higher N50 average length of Oxford Nanopore reads helps to generate a better contigous assembly in this case. Oxford Nanopore assembly requires substantial polishing with both long and Illumina short reads, however, the resulting assemblies are more contigous and easy to annotate. In future, additional long read coverage data should help resolve rest of Chr Y sequence.
Get in touch with us for a free consultation about your genome assembly or bioinformatics projects: sales@1010genome.com
Reference
Helen Skaletsky et al, The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature volume 423, pages 825–837 (2003)

