doi: https://doi.org/10.1101/847855
Link: https://www.biorxiv.org/content/biorxiv/early/2019/11/19/847855.full.pdf
Advancement of high throughput sequencing technologies have enabled researchers to identify or screen cancer causing structural variants. Long-read, single molecule sequencing technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have been shown as most reliable to identify SVs with substantial improvements to both sensitivity and specificity.
A recent study compares various sequencing platforms for identification of structural variations in breast cancer genome. Author applied whole genome sequencing technique and showed comparative analysis results using Illumina, PacBio and Oxford Nanopore sequencing platforms.
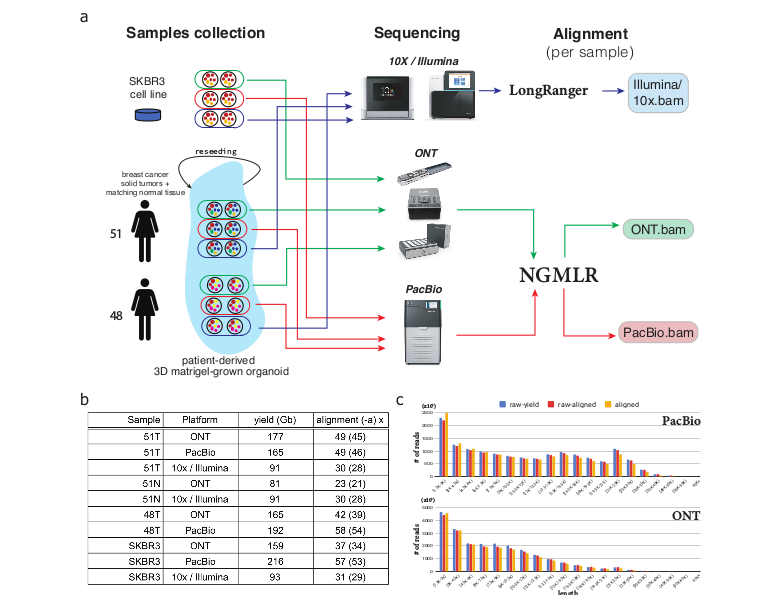
Sample collection, sequencing, and alignment pipeline and statistics overview
Sample Identification and sequencing
Tumor tissue samples were obtained from two affected females. Extracted DNA was used for parallel sequencing using ONT, PacBio and Illumina platforms.
Comparative analysis
For ONT and PacBio data-sets author used two state-of the art methods Sniffles and PBSV and for Illumina/10X data-set author used SV inference methods, with Lumpy, Manta, and SvABA designed for regular paired-end short Illumina reads, NAIBR, GrocSVS, and LongRanger which also utilize the single-molecule 10X Genomics barcode information.
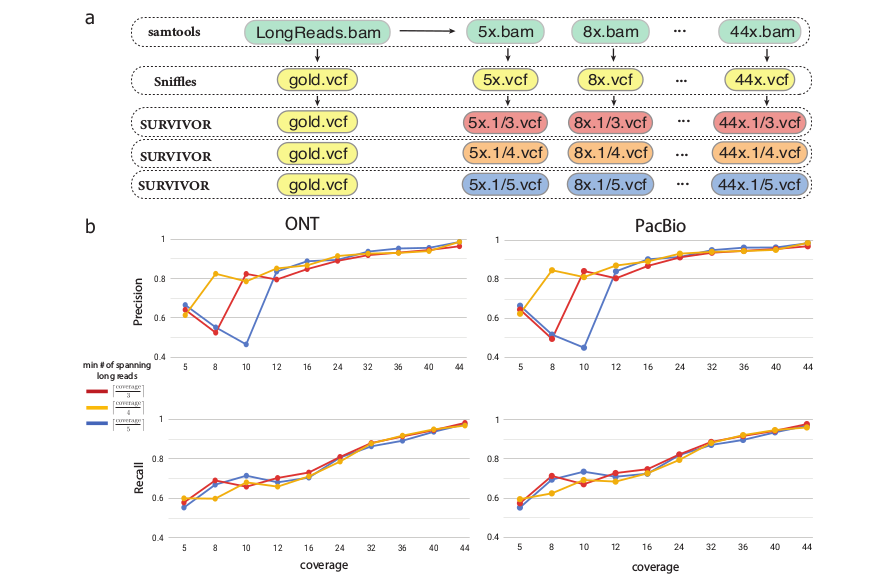
Structural variations inference on downsampled long-read datasets.
a) Workflow for downsampling full long-read dataset, and computing concordance between downsampled and full coverage datasets with distinct minimum fractional x/y read support for an SV to be considered.
b) Precision and Recall for SVs inferred on downsampled Oxford Nanopore and PacBio data for sample 51T. SVs inferred on the full coverage dataset at the matching read support threshold are used as the ground truth.
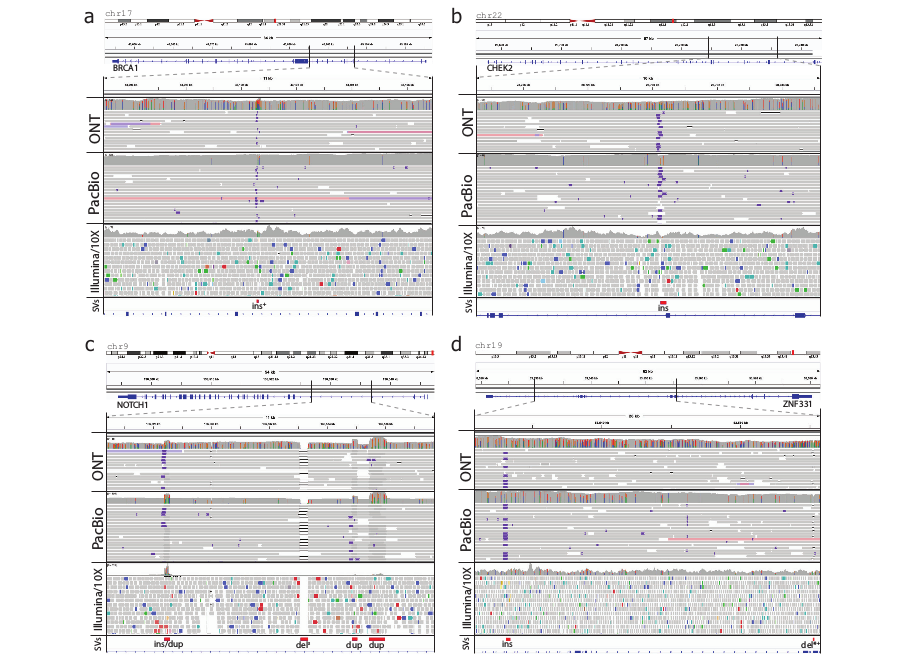
Above image shows SVs identified in cancer-related COSMIC census genes in patient 51. All presented SVs are identified with both Oxford Nanopore and PacBio reads.
Both Oxford Nanopore and PacBio identified 2 insertions in BRCA1 and CHEK2 breast cancer genes that were missed by Illumina short reads
In addition author also reported multiple SVs present in NOTCH1 and ZNF331 COSMIC census genes that have been known to be associated with tumor development.
If you are interested in getting your exome data analyzed or need help with exome projects, contact us for a no-obligation consultation at sales@1010genome.com or fill up the form below to provide us some details and one of our experts will contact you.

