Citation: 16 September 2019 / Accepted: 27 September 2019 / Published: 11 October 2019
Link: https://www.mdpi.com/2304-8158/8/10/480/htm
In recent years advancement in high throughput next generation sequencing has enabled researchers to detect the entire bacterial diversity/biodiversity spectra and foodborne pathogens in food which further reveals control of foodborne illness.
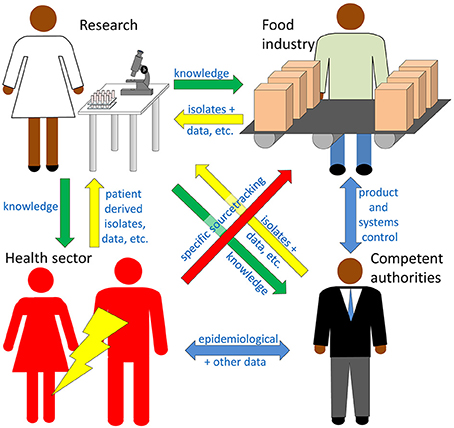
Image credit: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02029/full
Four sectors are considered here as potential users of high throughput sequencing (HTS) technologies for detection and characterization of foodborne pathogens.
16s rRNA gene amplicon sequencing for metagenome analysis
Three different commercial brands of RTE packed salads were analyzed in the present study. These salads contained radicchio (Cichorium intybus var. foliosum), arugula (Eruca sativa), foliosum), arugula (Eruca sativa), and lamb ́s lettuce (Valerianella locusta).
In first step as microbial culture total aerobic viable microorganisms (TVC), total coliforms, Escherichia coli, and Salmonella spp. were assessed to create a pool of microbial community which was to be referred with next generation sequencing.
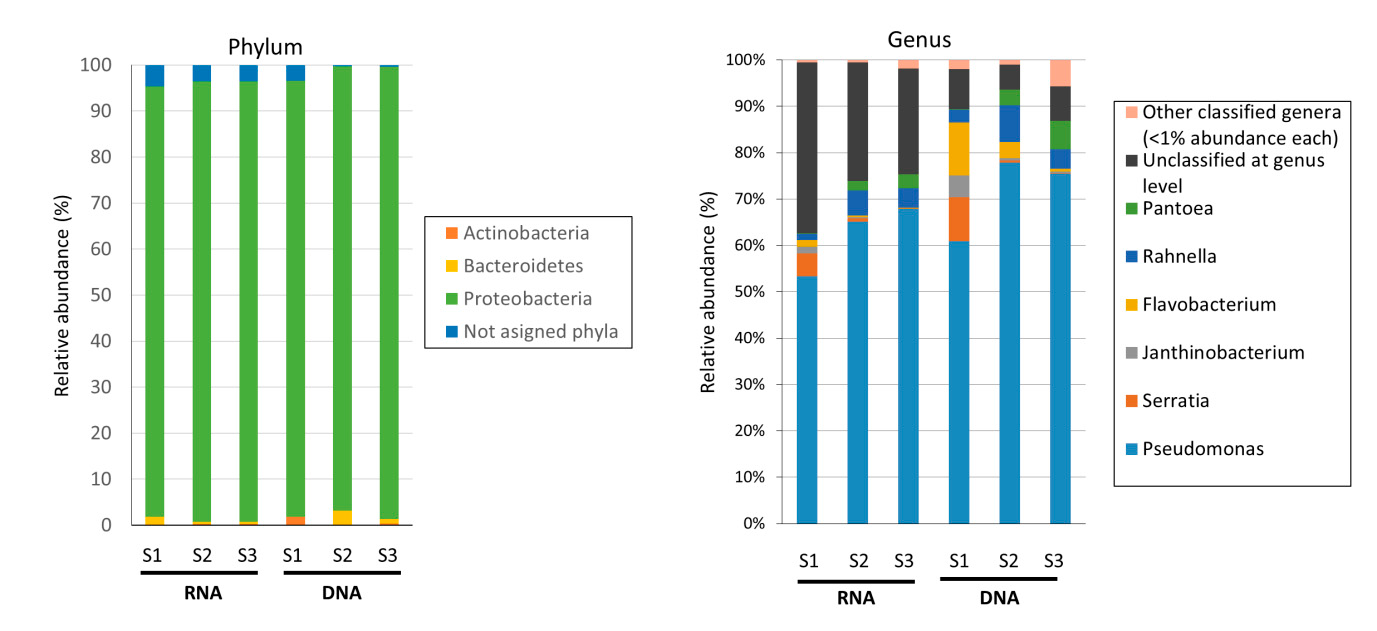
In second step high throughput Illumina next generation sequencing approach for 16s rRNA amplicon sequencing was performed from each one of the analyzed brands, since culture-based data pointed to very similar results, one replicate sample from each was used for high-throughput 16S rRNA sequencing of amplicons. Data analysis was performed using QIIME2 pipeline an operational taxonomic unit (OTU) threshold was set to 99% of nucleotide identity, with between 7201 and 12,848 OTUs obtained from sequenced samples.
High-throughput sequencing data of 16S rDNA and 16srRNA from RTE salads. Those low abundant genera that showed <1% of relative abundance were grouped within the category of “Other classified genera.

Principal component analysis (PcoA) of bacterial community structure at the genus level from the analysed RTE salads. This plot shows samples from total bacterial cells (dormant, dead, and active cells containing 16S rDNA genes) and metabolically active bacteria (cells containing ribosomal RNA, 16S rRNA) are depicted in red and blue colors, respectively. Two close samples in the PcoA indicated that they had similar bacterial species composition and relative abundances of taxa.
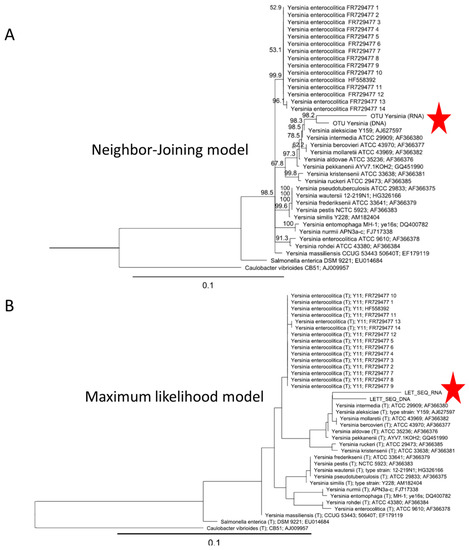
Phylogenetic tree of operational taxonomic units (OTUs) assigned to Yersiniaspp. Yersinia OTU confirmed that it was related to non-pathogenic Yersinia strains
This study re-emphasizes importance of 16s rRNA sequencing and metagenome analysis to address a important question; Which part of the bacterial community is active in a food sample?
High throughput Next generation sequencing approach of 16S rRNA and rDNA provides a more complete and robust view of the bacterial biodiversity and the active bacterial fraction present in food samples.
If you are interested in getting your 16s rRNA data analyzed or need help with 16s metagenome projects, contact us for a no-obligation consultation at sales@1010genome.com or fill up the form below to provide us with some details and one of our experts will contact you.

