Illumina Sequencing
Illumina sequencing or also known as Sequencing by Synthesis (SBS) sequencing is the most popular next-generation technology. About 90% of the sequencing is performed on illumina sequencing platforms. The speed, accuracy and cost effectiveness of illumina sequencing makes it the most popular choice for genomics community.
Why Illumina Sequencing Is An Standout?
Illumina sequencing is based on the “sequencing-by-synthesis” (SBS) technique where a modified DNA polymerase incorporates fluorescently labeled dNTPs into a DNA template during controlled cycles of DNA synthesis. The dNTPs are specially designed bases that have a fluorescent reversible terminator that allows addition of a single base at a time. Then this terminator is cleaved off it allows for additional of another dNTP while at the same time fluorophore on the cleaved terminator is excited by a laser beam. Though Illumina’s camera are unable to detect a single fluorescence event, a bridge amplification method allows to grow a lawn of millions of DNA template at a given location of a glass flow cell amplifying the signal in each cycle of DNA synthesis.

Illumina Sequencing Workflow has 3 Key Steps:
1. Library Preparation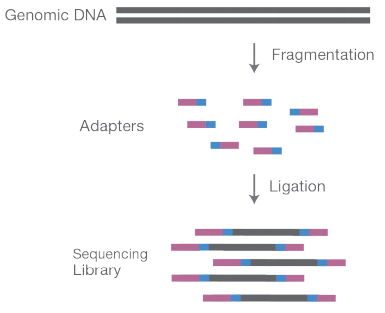
Genomics DNA is randomly fragmented into shorter fragments of ~200 bps followed by 5’ and 3’ end adapter ligations. These short fragments are then PCR amplified and purified.
2. Cluster Generation
Amplified library is loaded on to a flow cell where a high density lawn of immobilized oligos complementary to adapters capture short fragments of DNA. Bridge amplification process generates clonal clusters as shown in the figure below ready for sequencing.
3. Sequencing
A highly accurate base by base sequencing is performed to generate raw bases. Sophisticated base calling algorithms decipher the fluorescence signals from each cluster to output raw base calls and quality scores. The reversible terminator based SBS technique is so accurate that it eliminates any context bias caused by homopolymeric or repetitive regions in a genome resulting into a high raw base accuracy of ~99%.
Advantages of Illumina Sequencing
- Proven Base Calling Accuracy

Each run of illumina sequencing generates about 85% of bases above the quality score of Q30. This quality score in turn suggests that there is only a 0.1% likelihood of incorrect base calling. This accuracy make illumina sequencing most accurate with least false positive, false negative and missed call rates among all next-generation sequencing technology.
- Cost Effective
Massive parallel sequencing and clever optics makes illumina sequencing the most cost effective next-generation sequencing technology for most of the applications. Illumina sequencing has enabled to achieve a $1000 per human genome milestone with its latest novaseq instruments.
- High Quality Pairwise Alignments
Pair end illumina sequencing helps to improve sequence alignment for SNP detection, genome assemblies and structural variation detection like gene fusion, translocation and deletion events. Pair end libraries have a known distance between the read pair providing the alignment accuracy. Similarly, mate pair libraries of size 5kb, 10 and 20kb are very helpful for genome assembly and SV detection.
- Unbiased Genome Coverage
Latest improvement in SBS chemistry enable an unbiased and uniform coverage across difficult to sequence regions like GC-rich regions or repetitive regions.
Illumina Library Preparation Protocols
Pair End Sequencing
Paired end sequencing allows sequencing of both ends of a fragment and generate high-quality reads. As the distance between the 2 reads of a pair is known, pair end sequencing helps generate quality alignments to a genome to detect genomic rearrangements and repetitive sequence elements, as well as gene fusions and novel transcripts. Paired end library prep is a straight forward protocol that generation of unique ranges of insert sizes. This protocol is equally effective for sequencing gDNA or cDNA and methylation studies.
Single-Read Sequencing
Single-read sequencing involves sequencing DNA from only one end and is the simplest form of Illumina sequencing. This library preparation requires only 100 ng of gDNA or cDNA. Single end reads are a good alternative for Sanger sequencing and cost as low as a hundredth.
Mate-pair Sequencing
Mate pair protocol uses large fragments of size 2kb – 20kb. These large fragments are end-biotinylated to tag the partners followed by circularization and refragmentation that generated small size fragments. The fragments that has both mate pair ends without any intervening sequence are enriched by biotin tags to be sequenced just like normal pair-ends. The key advantage here is that the distance between the two ends of mate pair is known depending on the size of fragments generated at first step – 2kb to 20 kb. These mate pair reads when aligned to a reference genome should align at a distance equal to fragment size.
Usually, mate pair reads are combined with pair-end reads to generate deep coverage and accurate mapping. Such long-insert mate pair libraries are useful for a number of sequencing applications, including:
- De novo sequencing
- Genome finishing
- Structural variant detection
- Identification of complex genomic rearrangements
Details on Illumina sequencing library preparation protocols can be found in the genomics datasheet document at this link.
Applications of Illumina Sequencing
Illumina sequencing provides a wide range of tools to probe any given genome, transcriptome or epigenome. Most applications of illumina sequencing methods differ in the ways of gDNA or RNA extraction and library preparation methods while the process of sequencing remains the same. There are a number of library preparation kits available from Illumina and other vendors for a specific application. At 1010genome we have optimized sample extraction methods based on a specific application and organism type to deliver high quality illumina sequencing reads.
These are the key applications areas for Illumina Sequencing:
Unlock the power of your Illumina sequencing data with our expert analysis services.
We are committed to providing high-quality, cost-effective, and time-efficient services. We work closely with our clients to understand their specific needs and goals, and we deliver results that meet or exceed their expectations. Contact us today to learn more about how we can help you analyze your Illumina sequencing data.